介绍
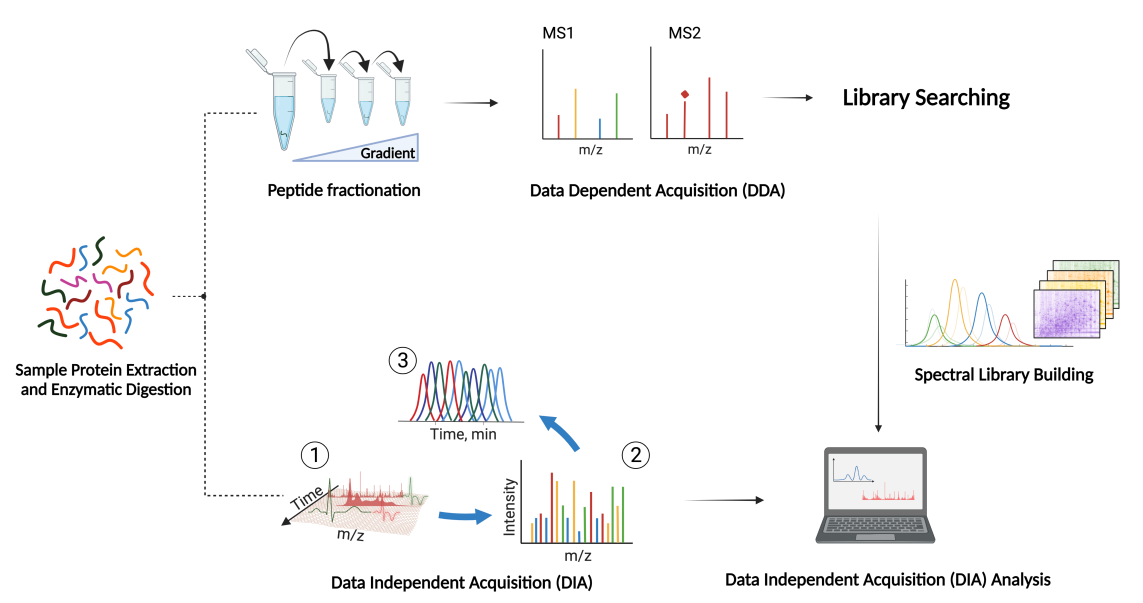
DIA(Data-Independent Acquisition,数据非依赖性采集)是一种无歧视性和无随机性的蛋白质组分析技术, 将质谱全扫描范围分为若干个窗口,然后对每个窗口中的所有离子进行检测及碎裂,从而无遗漏无差异地获得样本中所有离子的信息,降低样本检测的缺失值,同时提高定量准确性和重复性,实现大样本队列中高稳定,高精准的蛋白质组定量分析。
欧米服务
- 1. 西湖欧米结合 PCT 样本前处理技术,可以实现临床微量样本 (如 FFPE、穿刺活检、泪液等)的高深度蛋白质定量分析,组织样本最低送样量只需 0.1 mg。
- 2. 使用多种蛋白质组学搜库软件,包括 OpenSWATH、EncyclopeDIA、DIA-NN 等,并能够综合分析结果,提高蛋白的鉴定量和定量准确度。
- 3. 开发了优化特异性谱图库的方法,发明专利:基于优化数据库(Sub-Lib)的数据非依赖性质谱检测方法,专利号:202010773114.5。
质谱仪
Orbitrap Exploris 480
合作项目案例
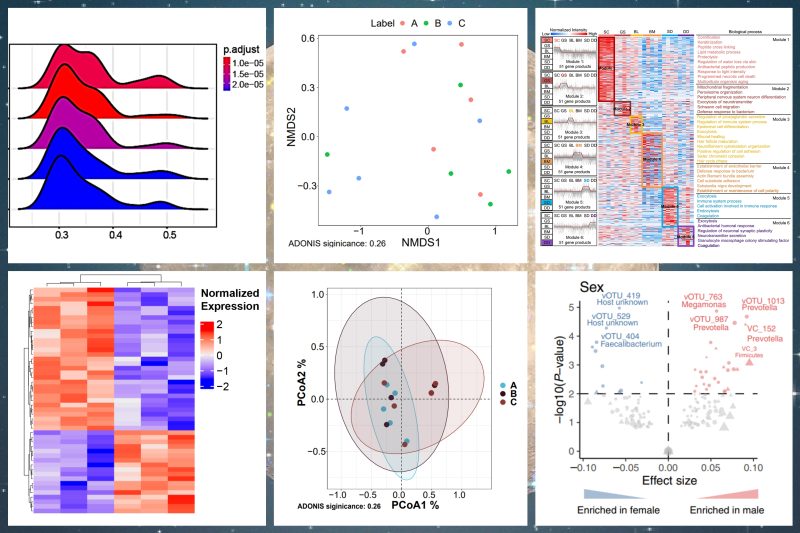
Cancer Communications | DIA 技术解析结直肠癌肿瘤组织 FFPE 样本蛋白组特征,揭示 PLOD2 是结直肠癌潜在治疗新靶点
该研究利用 PCT 微量样本前处理技术和 DIA 技术结合,在结直肠癌患者的 170 个 FFPE 组织样本中量化了 6359 种蛋白质,技术重复之间的相关系数为 0.953。该研究在不同临床阶段结直肠癌患者肿瘤组织样本和正常结肠组织样本中分析出表达水平显著差异的 928 种蛋白质,并进一步通过靶向表达水平验证和细胞水平的功能验证明确了 PLOD2 为潜在的 CRC 治疗靶点。
参考文献
2. Röst et al. OpenMS: a flexible open-source software platform for mass spectrometry data analysis. Nature Methods. 2016.13(9):741-748
https://www.nature.com/articles/nmeth.3959
3.Röst, et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nature Biotechnology. 2014.32:219-223
https://www.nature.com/articles/nbt.2841
4.Guo, et al. Rapid mass spectrometric conversion of tissue biopsy samples into permanent quantitative digital proteome maps. Nature Medicine. 2015.21(4):407–413.
https://www.nature.com/articles/nm.3807
5. Searle et al. Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry. Nature Communications. 2018. 9(1): 1-12
https://www.nature.com/articles/s41467-018-07454-w
6.Xu, et al. In-depth Serum Proteomics Reveals Biomarkers of Psoriasis Severity and Response to Traditional Chinese Medicine. Theranostics. 2019.9(9): 2475-2488.
https://www.thno.org/v09p2475.htm
7.Shao, et al. Comparative analysis of mRNA and protein degradation in prostate tissues indicates high stability of proteins. Nature Communications. 2019. 10(1):2524.
https://www.nature.com/articles/s41467-019-10513-5
8.Zhu, et al. High-throughput Proteomic analysis of FFPE tissue samples facilitates tumor stratification. Molecular Oncology. 2019 Sep;13(11): 2305-2328.
https://febs.onlinelibrary.wiley.com/doi/10.1002/1878-0261.12570
9.Zhang, et al. Data-Independent Acquisition Mass Spectrometry-Based Proteomics and Software Tools: A Glimpse in 2020. Proteomics. 2020.20(17-18): e1900276.
https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/pmic.201900276
10.Demichev, et al. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nature Methods. 2020.17:41-44
https://www.nature.com/articles/s41592-019-0638-x
11.Cai, et al. PulseDIA: Data-Independent Acquisition Mass Spectrometry Using Multi-Injection Pulsed Gas-Phase Fractionation. Journal of Proteome Research. 2021.20(1):279-288.
https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00381
12.Ge, et al. Computational Optimization of Spectral Library Size Improves DIA-MS Proteome Coverage and Applications to 15 Tumors. Journal of Proteome Research. 2021
https://pubs.acs.org/doi/full/10.1021/acs.jproteome.1c00640
13.Liu, et al. DIA-based Proteomics Identifies IDH2 as a Targetable Regulator of Acquired Drug Resistance in Chronic Myeloid Leukemia. Mol Cell Prot. available at bioRxiv, 2021.
https://www.mcponline.org/article/S1535-9476(21)00159-6/fulltext#secsectitle0030
14.Shao, et al. Proteomics profiling of colorectal cancer progression identifies PLOD2 as a potential therapeutic target. Cancer Commun. 2021.
https://onlinelibrary.wiley.com/doi/10.1002/cac2.12240
15.Zhu, et al. Snapshot: Clinical proteomics. Cell. 2021.184(18): 4840-4840.