Introduction
TMT (Tandem Mass Tag) technology is a peptide labeling technique developed by Thermo Scientific, Inc. Westlake Omics uses the latest TMTpro 16plex isotope labeling reagent set to perform tandem mass spectrometry analysis by specifically labeling specific amino groups of peptides, allowing the simultaneous relative quantification of proteins of up to 16 different samples. TMT-labeled peptides are typically acquired in DDA (data-dependent acquisition) mode (labeled DDA).
Service
1. Westlakeomics uses TMTpro 16plex isotope labeling reagent group can achieve simultaneous comparative analysis of relative protein content among 500 samples through design optimization and batch effect processing.
2. The quantitative quantity of protein in tissue samples can reach 10000+ and that in blood can reach 1000+ through 30 Fraction.
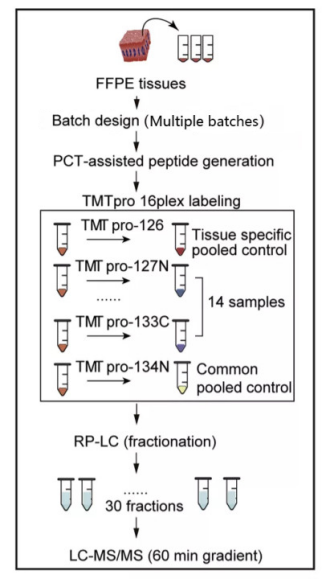
3. Combined with PCT sample pre-processing technology, it can achieve high depth protein quantitative analysis of clinical micro samples (such as FFPE, puncture biopsy, tears, etc.), and the minimum sample size of tissue sample is only 0.1mg.
Mass Spectrometer
Orbitrap Exploris 480
Project Case
FFPE samples have always been a very important clinical sample resource, but it is difficult to carry out proteomics research because of its small sample size. In this study, 11394 proteins were quantified in 218 FFPE samples from 7 different organs and tissues of 19 COVID-19 patients and 56 non COVID-19 patients infected with other diseases through surgery by PCT micro sample pre-processing technology and TMT labeling quantitative technology.
Case 2:Cell discovery | Proteomic characterization of Omicron SARS-CoV-2 host response
In this study, a total of 1464 proteins were quantified in 79 blood samples by depth grading TMT labeled quantitative proteomics, and 1155 proteins were left after the deletion value was filtered. 751 of these proteins were functionally analyzed, and the blood proteome characteristics of non critical patients infected with the Omicron variant were evaluated, as well as the molecular characteristics of the protective effect of the new crown vaccine on the infection of the Omicron variant.
References
1. Li, et al, TMTpro reagents: a set of isobaric labeling mass tags enables simultaneous proteome-wide measurements across 16 samples. Nature Methods. 2020. (17): 399-404.
2. Shen, et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell. 2020. 182(1): 59-72.
3. Nie, et al. Multi-organ Proteomic Landscape of COVID-19 Autopsies. Cell. 2021 Feb.184(3): 775-791.
4. Zhu, et al. Snapshot: Clinical proteomics. Cell. 2021.184(18): 4840-4840.
5. Bi, et al. Proteomic and metabolomic profiling of urine uncovers immune responses in COVID-19. Cell Reports. 2021. 38(3)